Advancements in whole genome sequencing have ignited a revolution in digital biology. As the cost of high-throughput, next-generation sequencing has declined, genomics programs across the world are gaining momentum.
Whether used for sequencing critical-care patients with rare diseases or in population-scale genetics research, whole genome sequencing is becoming a fundamental step in clinical workflows and drug discovery.
But genome sequencing is just the first step. Analysing genome sequencing data requires accelerated compute, data science and AI to read and understand the genome.
Sequencing an individual’s whole genome generates roughly 100Gb of raw data. That more than doubles after the genome is sequenced using complex algorithms and applications such as deep learning and natural language processing.
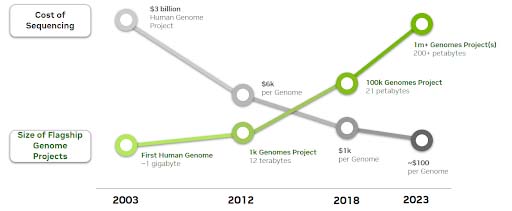
As the cost of sequencing a human genome continues to decrease, volumes of sequencing data are exponentially increasing.
An estimated 40 exabytes will be required to store all human genome data by 2025. As a reference, that’s 8-times more storage than would be required to store every word spoken in history.
Many genome analysis pipelines are struggling to keep up with the expansive levels of raw data being generated.
Sequencing analysis is complicated and computationally intensive, with numerous steps required to identify genetic variants in a human genome.
Deep learning is becoming important for base calling right within the genomic instrument using RNN- and convolutional neural network (CNN)-based models. Neural networks interpret image and signal data generated by instruments and infer the 3-billion nucleotide pairs of the human genome.
This is improving the accuracy of the reads and ensuring that base calling occurs closer to real time, further hastening the entire genomics workflow, from sample to variant call format to final report.
For secondary genomic analysis, alignment technologies use a reference genome to assist with piecing a genome back together after the sequencing of DNA fragments.
BWA-MEM, an algorithm for alignment, is helping researchers rapidly map DNA sequence reads to a reference genome. STAR is another alignment algorithm used for RNA-seq data that delivers accurate, ultrafast alignment to better understand gene expressions.
The programming algorithm Smith-Waterman is also widely used for alignment, a step that’s accelerated 35-time on the Nvidia H100 Tensor Core GPU, which includes a dynamic programming accelerator.
Uncovering genetic variants
One of the most critical stages of sequencing projects is variant calling, where researchers identify differences between a patient’s sample and the reference genome. This helps clinicians determine what genetic disease a critically ill patient might have, or helps researchers look across a population to discover new drug targets.
These variants can be single-nucleotide changes, small insertions and deletions, or complex rearrangements.
GPU-optimised and -accelerated callers such as the Broad Institute’s GATK – a genome analysis toolkit for germline variant calling – increase speed of analysis.
To help researchers remove false positives in GATK results, Nvidia collaborated with the Broad Institute to introduce NVScoreVariants, a deep learning tool for filtering variants using CNNs.
Deep learning-based variant callers such as Google’s DeepVariant increase accuracy of calls, without the need for a separate filtering step. DeepVariant uses a CNN architecture to call variants. It can be retrained to fine-tune for enhanced accuracy with each genomic platform’s outputs.
Secondary analysis software in the Nvidia Clara Parabricks suite of tools has accelerated these variant callers up to 80t-times. For example, germline HaplotypeCaller’s runtime is reduced from 16 hours in a CPU-based environment to less than five minutes with GPU-accelerated Clara Parabricks.
Accelerating the next wave of genomics
Nvidia is helping to enable the next wave of genomics by powering both short- and long-read sequencing platforms with accelerated AI base calling and variant calling. Industry leaders and startups are working with Nvidia to push the boundaries of whole genome sequencing.
For example, biotech company PacBio recently announced the Revio system, a new long-read sequencing system featuring Nvidia Tensor Core GPUs. Enabled by a 20-times increase in computing power relative to prior systems, Revio is designed to sequence human genomes with high-accuracy long reads at scale for under $1 000.
Oxford Nanopore Technologies offers the only single technology that can sequence any-length DNA or RNA fragments in real time. These features allow the rapid discovery of more genetic variation. Seattle Children’s Hospital recently used the high-throughput nanopore sequencing instrument PromethION to understand a genetic disorder in the first few hours of a newborn’s life.
Ultima Genomics is offering high-throughput whole genome sequencing at just $100 per sample, and Singular Genomics’ G4 is the most powerful benchtop system.

